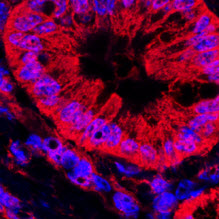
Research Projects
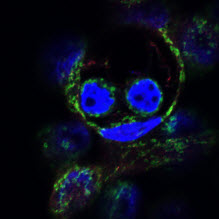
Mitochondria are often pictured in biology texts as static, individual organelles, when in reality they form highly dynamic and mobile networks constantly undergoing fusion and fission. Research in the last decade has revealed that mitochondrial fusion and fission impact nearly every aspect of mitochondrial function, from cellular metabolism, to calcium homeostasis, to the control of cell survival through apoptosis and autophagy. In addition, failure to properly maintain this dynamic mitochondrial network has been shown to play a role in a number of diseases, including Parkinson’s disease, Alzheimer’s disease, diabetes and cancer. The goals of our research are: (1) to uncover the molecular mechanisms that directly control mitochondrial dynamics, (2) to identify and characterize signaling pathways that interact with the mitochondrial dynamics machinery and (3) to determine how these interactions impact the pathology of diseases such as cancer.
1. Mitochondrial dynamics in cancer
Mitochondrial dynamics have been shown to be important for the cellular control of apoptosis, autophagy and metabolic function, processes that are critical regulators of tumorigenesis. As such, we seek to understand the molecular basis of how oncogenic signaling pathways converge upon the mitochondrial fusion and fission machinery and how mitochondrial dynamics contribute to oncogenic transformation and tumorigenesis. To that end, we use a combination of patient-derived xenografts and genetically engineered mouse models to determine the role that mitochondria shaping proteins play in the initiation and maintenance of human tumors.
2. Pathways that regulate mitochondrial dynamics
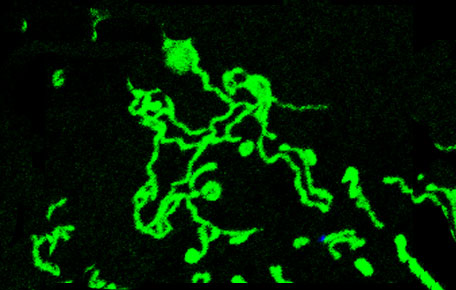 A number of important cellular processes are accompanied by changes in the mitochondrial network, but the signaling pathways that link these processes to the mitochondrial fusion and fission machinery are not well understood. Using unbiased approaches such as large-scale proteomic and RNAi screens, as well as more targeted gain- and loss-of-function analyses, we are identifying and characterizing signaling pathways that regulate mitochondrial fusion and fission. To facilitate this approach, we develop novel computational tools that enable high throughput analysis of mitochondrial morphology in both cell lines and tissue. Furthermore, using a combination of biochemistry, molecular genetics and cell biology, we investigate how these pathways interact with the core mitochondrial fusion and fission machinery.
A number of important cellular processes are accompanied by changes in the mitochondrial network, but the signaling pathways that link these processes to the mitochondrial fusion and fission machinery are not well understood. Using unbiased approaches such as large-scale proteomic and RNAi screens, as well as more targeted gain- and loss-of-function analyses, we are identifying and characterizing signaling pathways that regulate mitochondrial fusion and fission. To facilitate this approach, we develop novel computational tools that enable high throughput analysis of mitochondrial morphology in both cell lines and tissue. Furthermore, using a combination of biochemistry, molecular genetics and cell biology, we investigate how these pathways interact with the core mitochondrial fusion and fission machinery.
3. The regulation of organelle dynamics by cellular stress
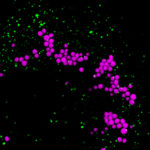 Cells in the body regularly encounter stress, and a number of pathways have evolved that allow cells to maintain homeostasis under a broad range of conditions. For example, under low nutrient conditions, cells adapt in part by recycling cellular lipids and storing them in lipid droplets in the form of triglycerides. Eventually, these lipid stores can be transferred to the reticular mitochondrial network to be used as fuel. We are currently exploring the mechanisms through which the cell coordinates this cellular response. In particular, we are investigating how multiple organelles, including the ER, lysosomes, lipid droplets and mitochondria communicate with each other to facilitate efficient recycling of cellular lipids. In addition, we are employing both GEMM and PDX tumor models to understand how this stress response contributes to tumor cell dormancy and metastasis.
Cells in the body regularly encounter stress, and a number of pathways have evolved that allow cells to maintain homeostasis under a broad range of conditions. For example, under low nutrient conditions, cells adapt in part by recycling cellular lipids and storing them in lipid droplets in the form of triglycerides. Eventually, these lipid stores can be transferred to the reticular mitochondrial network to be used as fuel. We are currently exploring the mechanisms through which the cell coordinates this cellular response. In particular, we are investigating how multiple organelles, including the ER, lysosomes, lipid droplets and mitochondria communicate with each other to facilitate efficient recycling of cellular lipids. In addition, we are employing both GEMM and PDX tumor models to understand how this stress response contributes to tumor cell dormancy and metastasis.